Mitochondria are essential for eukaryotic life due to their critical role in central metabolic pathways. Owing to the close proximity to respiratory chain system, which is the site of mitochondrial reactive oxygen species (ROS) production, mitochondrial DNA (mtDNA) suffers more oxidative damage than nuclear DNA. Since the discovery that unrepaired lesions lead to the instability of mitochondrial genome, which associate with aging and a wide variety of severe human diseases, the mechanisms of mtDNA repair have been studied extensively. Still, DNA repair in mitochondria has not been fully elucidated and many fundamental questions remain unanswered.
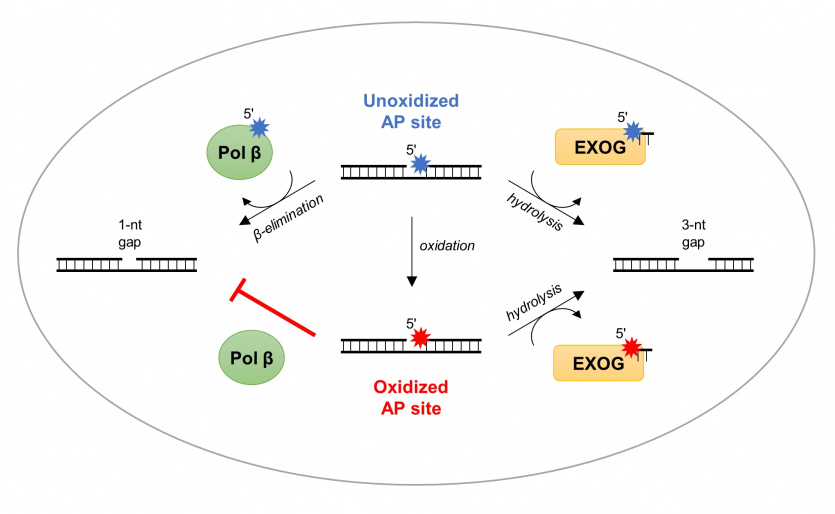
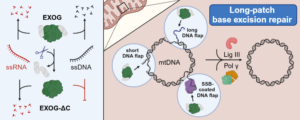
Most oxidative damage in mitochondrial DNA is resolved by the base excision repair (BER), a multi-enzymatic reaction pathway. However, the enzyme that catalyzes the rate-limiting reaction – deoxyribose phosphate (dRP) moiety removal – has not been completely determined in mitochondria. 5’-dRP moiety is formed after the removal of a damaged base, thus creating abasic (AP) site, followed by DNA backbone incision. In our recent work we presented structural and enzymatic studies showing that human mitochondrial exonuclease EXOG exhibits strong dRP removal. Unlike canonical dRP lyases (e.g. repair polymerase Pol β in nucleus) that act only on intact dRP, EXOG processes a variety of abasic sites, including dRP, its oxidized product deoxyribonolactone (dL) and the stable synthetic analog tetrahydrofuran (THF). We determined crystal structures of EXOG complexed with THF-containing DNA and with a partially-gapped DNA, which, together with biochemical analyses, provided the mechanism for the EXOG-mediated processing of a broad spectrum of substrates. We showed that EXOG uses a controlled 5’-exonuclease activity to cleave the third phosphodiester bond away from the abasic site, thus providing independence from a 5’ damage type.
Through structural and enzymatic analyses in our work, we have shown that the human mitochondrial exonuclease EXOG has very potent dRP removal activity. Unlike canonical dRP lyases (e.g., Pol β repair polymerase in the cell nucleus), which remove only intact dRP, EXOG is capable of excising different AP sites: (1) dRP, (2) deoxyribonolactone dL – an oxidized form of dRP, and (3) tetrahydrofuran THF – a synthetic analog of dRP. We solved the crystal structure of EXOG in complex with THF-containing DNA, as well as with a partial nucleotide break.
The structural data obtained, together with biochemical analyses, allowed us to propose a mechanism for EXOG activity on DNA substrates containing different damage. We have shown that with its controlled 5′-egzonucleolytic activity, EXOG cuts the third phosphodiester bond from the AP site, allowing the removal of damage in mtDNA regardless of its type.
This work was created as part of the POLONEZ 2 grant of the National Science Center (NCN).